In our world, axolotls are known because of their resemblance to Pokémon. However, axolotls also exhibit traits that are unique to their species – what I like to call superpowers. They have the neoteny trait, through which they stay young forever, as well as the ability to self-heal. This is the reason why scientists have been thoroughly examining their DNA, where they believe they can find answers to support regenerative medicine.
Today, nearly 2 million people in the US alone experience limb loss. Latest therapeutic interventions show us a great advance in prosthetic limbs as a way of replacement. Prosthetics are a great way to improve people’s lives and restore mobility and independence. However, scientists are discovering unexplored regions. Is there a possibility for humans to regrow an amputated limb? To begin to answer this question, scientists have studied animals that have this ability already. The axolotl is an amphibian that can regenerate multiple structures like limbs, tail, spinal cord, skin and more without experiencing scarring in their lifetime. This animal has been cultivated in labs for more than 150 years thanks to this power and the simplicity in breeding.
Surprisingly, the axolotl’s genome is the largest one ever to be decoded, spanning about ten times the size of the human genome. Studying the genome is important to understanding biological processes, evolution, diseases and conservation. Scientists from the Research Institute of Molecular Pathology used an approach to study the axolot’s genome that combined “long-read sequencing, optical mapping and development of a new genome assembler (MARVEL).” Long-read sequencing, named the method of the year, is an innovative technique in which long DNA or RNA structures are read with great precision and with high efficiency. This is a more accurate method where scientists can figure out the gene family evolution. According to Erich Jarvis, a Rockefeller University researcher, this method “makes it possible to measure gene network interactions across chromosomes in ways not previously possible.” Optical mapping is a great complement to this technique, as it is in charge of the visual aspect of studying the DNA using imaging modalities.
Using the MARVEL approach, they discovered that the axolotl has the PAX7 gene but not the PAX3 (Nowoshilow et al., 2018). Paired genes (PAX genes) are a family of developmental and control genes that play an essential role in the formation of tissues and organs. For example, in vertebrates, Pax genes are involved in embryonic development. Recent research on spontaneous and transgenic mouse mutants has revealed that vertebrate Pax genes are key regulators during the formation of the kidney, eye, ear, nose, vertebral column and brain, possibly resulting in the determining the moment and location of organ morphogenesis (Dahl et al., 1997). In order to evaluate the functional consequences of the Pax 3’s absence in the axolotl, they employed TALEN- and CRISPR- mediated gene editing.
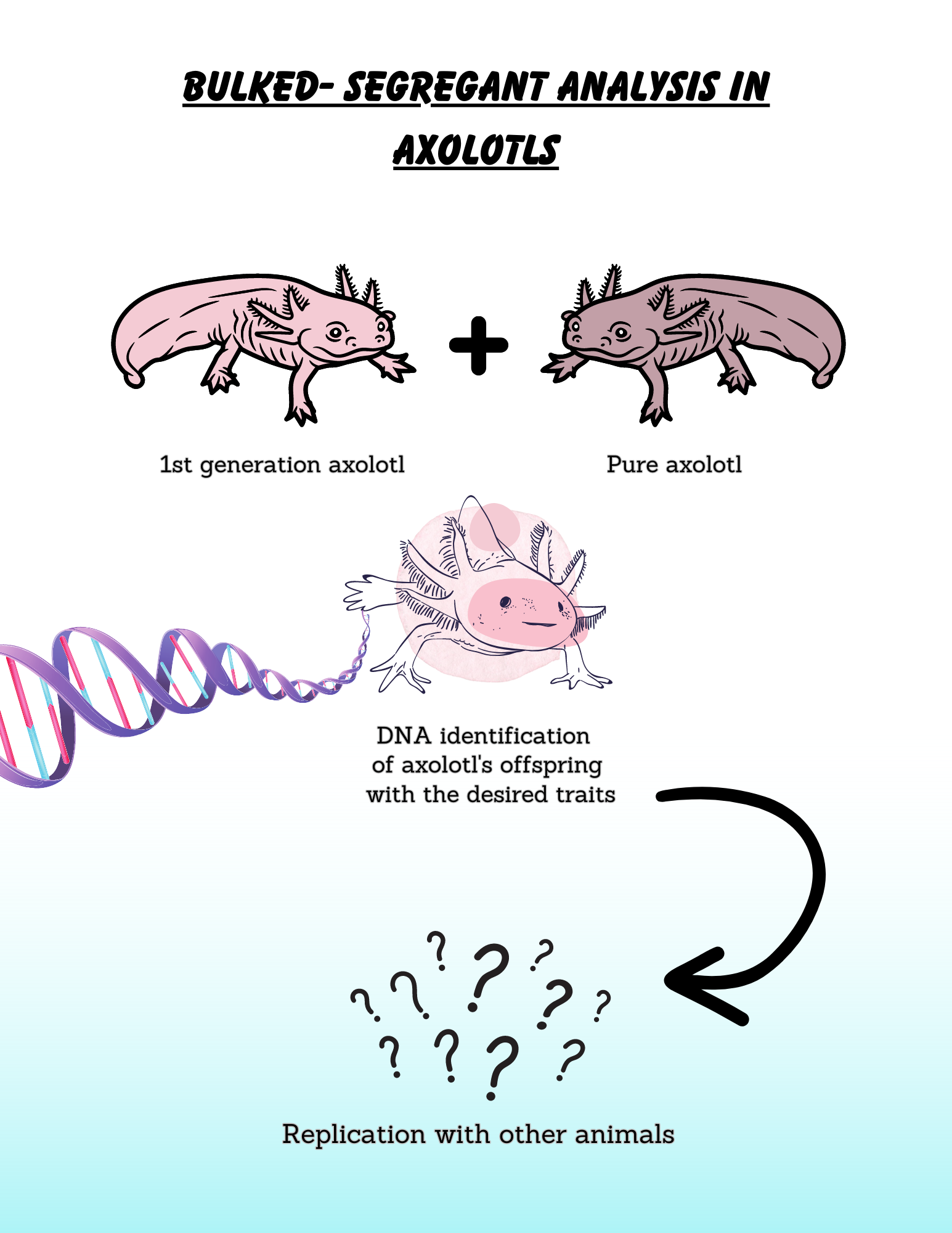
Based on previous research, in which scientists found that commonly studied axolotls derive from the tiger salamander (Ambystoma tigrinum mavortium), researchers decided to recross this first-generation axolotls with pure axolotls. DNA was extracted from 48 of these second generation individuals using a method called “bulked segregant analysis.” This is an efficacious method that is used to identify genes tied to specific attributes in living organisms. It starts when scientists contrast two groups or “bulks” of the same species: one with the desired attribute and the other one without it. The DNA is then scanned using genetic markers as they are SNPs (Single Nucleotide Polymorphisms) and SSRs (Simple Sequence Repeats). When there are SNPs, there is a single nucleotide substitution; while with SSRs, this is done with repeated sequences of nucleotides. In simple terms, specific genes are mapped to see if they are associated with a trait, implying a possible linkage. If scientists are able to identify common patterns between species, they might be able to extend the possibility to use the same method with human beings.
Now, knowing the way researchers are studying the axolotl’s genes for broadening the scope of regenerating limbs, what can this animal teach us about bone reparation? Researchers decided to compare bone healing in the axolotl and mouse. In the beginning, the development of the limb skeleton of both species is cartilaginous. However, axolotls don’t have a standardized fracture fixation. So, scientists presented a surgical technique to stabilize the surgical cutting of the bone with a fixator plate, comparing it to a non-stabilized cut, and to limb amputation. Femurs that were fixated with a plate regained bone integrity more efficiently in comparison to the ones without a plate. Healing of a non-critical bone cut was incomplete after 9 months. Nevertheless, amputated limbs efficiently restored bone length and structure. Three weeks after injury, there was an accumulation of PCNA proliferating cells, very similar to mice with plate-fixed fractures, and non-fixated fractures (Polikarpova et al., 2022). This discovery makes the axolotl more similar to the mouse than thought, allowing scientists to stay engaged in the examination of this fascinating amphibian.
In conclusion, the axolotl is an intriguing creature that has abilities that are currently being carefully explored. Limb regeneration, bone healing…you name it. With a variety of amazing traits, the axolotl is a hidden gem that is not being hidden anymore. Such knowledge could open the way for transformative treatments that might one day allow humans to imitate this regenerative potential.
References
Nowoshilow, S., Schloissnig, S., Fei, JF. et al. (2018). The axolotl genome and the evolution of key tissue formation regulators. Nature 554, 50–55. https://doi.org/10.1038/nature25458
Ziegler-Graham, K., MacKenzie, E. J., Ephraim, P. L., Travison, T. G., & Brookmeyer, R. (2008). Estimating the prevalence of limb loss in the United States: 2005 to 2050. Archives of physical medicine and rehabilitation, 89(3), 422–429.https://doi.org/10.1016/j.apmr.2007.11.005
Marx, V. (2023). Method of the year: long-read sequencing. Nat Methods 20, 6–11. https://doi.org/10.1038/s41592-022-01730-w
Manly, D. (2011, April 13). “Regeneration: The Axolotl Story.” Scientific American Blog Network, 13 Apr. 2011, blogs.scientificamerican.com/guest-blog/regeneration-the-axolotl-story/.
Timoshevskiy, V. A., Keinath, M. C., & Timoshevskaya, N. (2019, January). A chromosome-scale assembly of the axolotl genome - researchgate. ResearchGate. https://www.researchgate.net/publication/330604870_A_chromosome-scale_assembly_of_the_axolotl_genome
Dahl, E., Koseki, H., & Balling, R. (1997). Pax genes and organogenesis. BioEssays : news and reviews in molecular, cellular and developmental biology, 19(9), 755–765. https://doi.org/10.1002/bies.950190905
Mansouri A. (1998). The role of Pax3 and Pax7 in development and cancer. Critical reviews in oncogenesis, 9(2), 141–149. https://doi.org/10.1615/critrevoncog.v9.i2.40
Polikarpova, A., Ellinghaus, A., Schmidt-Bleek, O., et al. (2022, June 30). The specialist in regeneration-the axolotl-A suitable model to study bone healing?. Regen Med 7,35. https://www.nature.com/articles/s41536-022-00229-4
